| CNV Workshop: | A web-enabled platform for analyzing genome variation such as copy number variation (CNV). |
Features:
|
|
Find out More/Join Project
To find out more about CNV Workshop or to join the project, please visit the
project summary page.
Download
- See README.txt to find out what to download and how to install.
- After that, visit the file download area and download what you need.
Support
Active tickets
View active
tickets for this project.
Source code
Source code for this project is available through its Subversion repository.
Screen shots
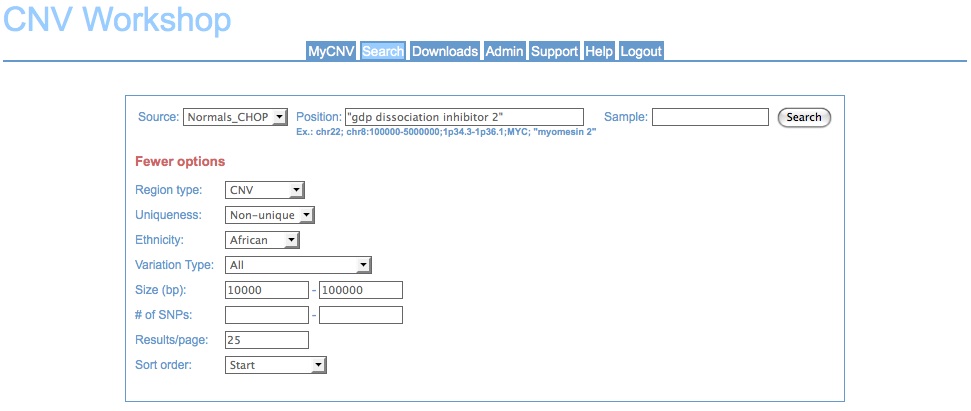
Search tab showing standard/basic search options for a user data set:
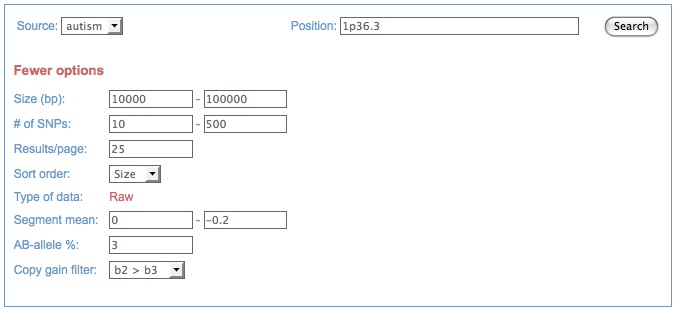
Search tab showing raw/advanced data search options for a user data set:
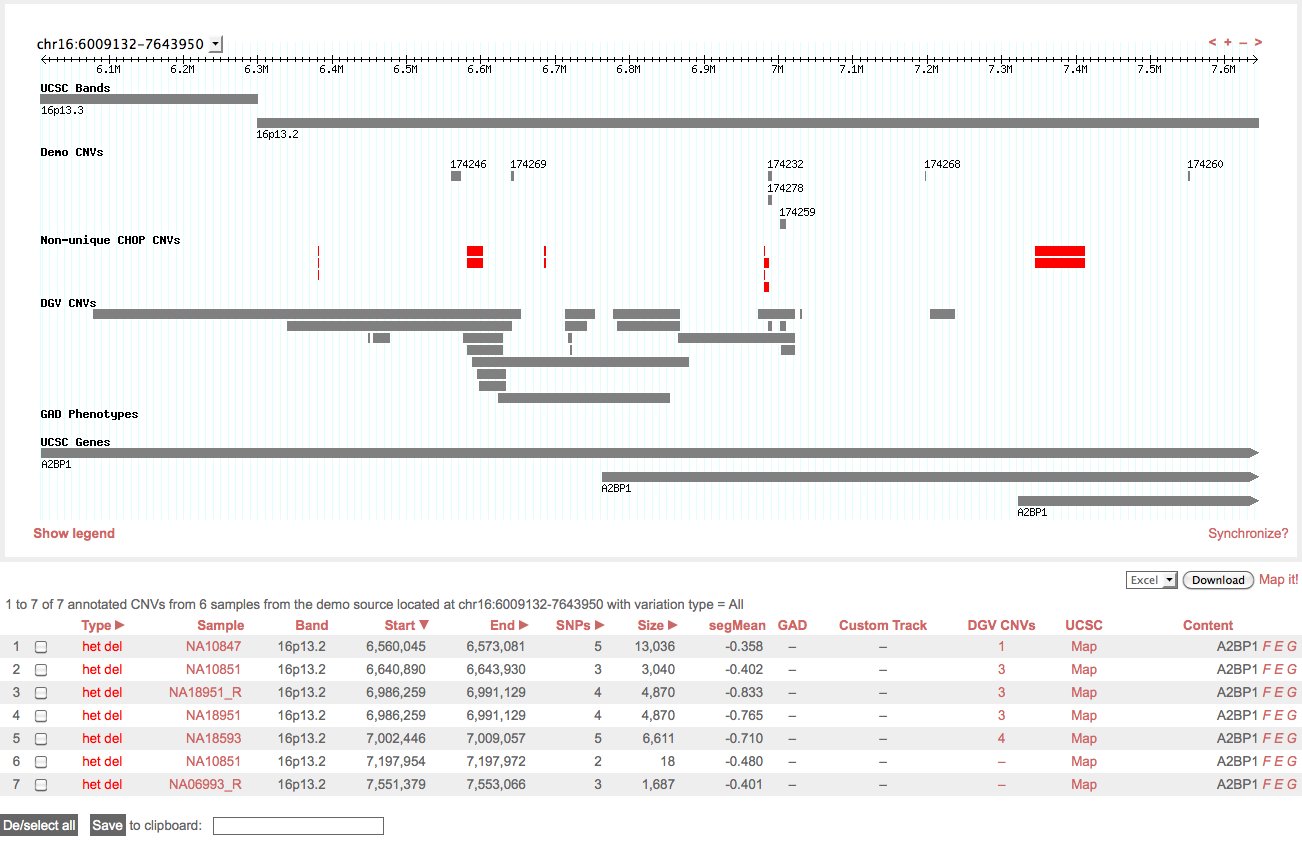
Search results page showing GBrowse visualization of a search region followed by tabular data: